Innovative Advances in Prenatal Screening Techniques
Written on
Chapter 1: The Rise of Non-Invasive Prenatal Testing
Recent developments in prenatal screening methods have sparked interest in their potential to predict fetal abnormalities through the analysis of Single Nucleotide Polymorphisms (SNPs). These tests are pivotal in identifying genetic predispositions to various birth defects.
This paragraph will result in an indented block of text, typically used for quoting other text.
Section 1.1 Overview of Traditional Screening Methods
Prenatal screenings serve to indicate whether a fetus may be at risk for certain genetic disorders. Common practices include the monitoring of maternal serum, ultrasounds, amniocentesis, and chorionic villus sampling (CVS). However, traditional serum detection methods when combined with ultrasound indicators such as nuchal translucency often carry false-negative rates ranging from 12% to 23%. The detection rates themselves can vary between 75% and 96%, with false-positive rates between 1.9% and 5.2%. Invasive techniques like amniocentesis and CVS carry risks, including infection, bleeding, and even miscarriage due to the nature of sample collection.
Section 1.2 The Emergence of NIPT
Non-invasive prenatal testing (NIPT) was introduced in 2011, utilizing cell-free fetal DNA (cffDNA) found in maternal plasma as a screening method for conditions like trisomy 21 (Down syndrome). Clinicians have embraced NIPT, particularly for women at higher risk for fetal aneuploidies, and it has been expanded to include low-risk populations in private practice. cffDNA originates from the syncytiotrophoblast layer of the placenta and has transformed the landscape of fetal screening, allowing for the detection of common aneuploidies such as trisomy 21, 18 (Edwards syndrome), and 13 (Patau syndrome). Additionally, this method can reveal fetal sex and identify sex chromosome aneuploidies, microdeletions, and monogenic disorders.
The first video titled "Prenatal screening methods" delves into various approaches for assessing fetal health, highlighting the advantages and limitations of each method.
Subsection 1.2.1 Advancements with Next Generation Sequencing
The advent of Next Generation Sequencing (NGS) has facilitated the integration of NIPT as a primary testing option in clinical settings. This technology accurately measures fetal fraction—the ratio of cffDNA to the total circulating cell-free DNA (cfDNA) in maternal plasma—thus influencing assay sensitivity. The average fetal fraction ranges between 10% and 15% during the 10 to 20 weeks of gestation. Previously, Y chromosome markers were utilized to ascertain fetal fraction, but current methods employ SNP ratios, which provide estimates independent of the fetus's sex.
Section 1.3 Innovations in SNP-Based Testing
The initial SNP-based NIPT methods relied on allele-specific PCR amplification of SNP markers derived from both paternal and maternal DNA. By comparing the intensities of maternal and fetal DNA bands, researchers could estimate the dosage of fetal chromosome 21, while unique paternal SNPs were used for fetal fraction estimation. Contemporary techniques leverage multiplex PCR of highly polymorphic SNPs followed by NGS to analyze single nucleotide variations and copy number variations.
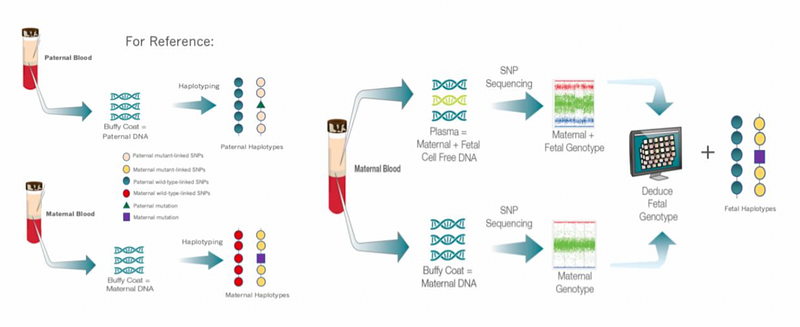
Figure 1. Overview of SNP-based Non-Invasive Screening Methods illustrates the collection of DNA samples from both parents and how these are analyzed to determine fetal inheritance patterns. Circles represent polymorphic alleles, triangles denote paternal mutations, and squares indicate maternal mutations. The methodology involves PCR amplification and sequencing to calculate risk scores for various chromosomes.
Chapter 2: Limitations and Future Directions
While SNP-based NIPT holds promise, it faces limitations, particularly in cases involving surrogacy or egg donation from unrelated donors. Moreover, the method's effectiveness can diminish in scenarios of high homozygosity between the mother and fetus, often due to consanguinity or uniparental disomy.
The second video, "Part 3: What is NIPT? (non-invasive prenatal testing)," provides a comprehensive overview of NIPT, discussing its methodologies and implications for prenatal care.
The non-invasive nature of NIPT makes it a highly recommended option for expectant parents. By differentiating between fetal and maternal DNA, this SNP-based approach proves advantageous in clinical settings, not only for identifying common aneuploidies but also for screening potential microdeletions. Furthermore, it opens avenues for cost-effective screening of other genetic disorders like beta-thalassemia and cystic fibrosis through the identification of paternal alleles. Innovative primer design for additional known disease SNP loci is on the horizon. The primary challenge remains the high incidence of false positives and inconclusive results due to low fetal fractions. However, advanced sequencing techniques, including deeper reads for high-risk assessments and refined risk calculations, are helping to address these issues. The ongoing development and refinement of SNP-based prenatal screening techniques are crucial in reducing unnecessary invasive procedures while improving detection rates for affected pregnancies.